Synthetic ion channels via self-assembly: a route for embedding porous polyoxometalate nanocapsules in lipid bilayer membranes
Porous polyoxometalate nanocapsules of Keplerate type are known to exhibit the functionality of biological ion channels; however, their use as an artificial ion channel is tempered by the high negative charge of the capsules, which renders their spontaneous incorporation into a lipid bilayer membrane unlikely. In this Letter we report coarse-grained molecular dynamics simulations that demonstrate a route for embedding negatively charged nanocapsules into lipid bilayer membranes via self-assembly. A homogeneous mixture of water, cationic detergent, and phospholipid was observed to spontaneously self-assemble around the nanocapsule into a layered, liposome-like structure, where the nanocapsule was enveloped by a layer of cationic detergent followed by a layer of phospholipid. Fusion of such a layered liposome with a lipid bilayer membrane was observed to embed the nanocapsule into the lipid bilayer. The resulting assembly was found to remain stable even after the surface of the capsule was exposed to electrolyte. In the latter conformation, water was observed to flow into and out of the capsule as Na(+) cations entered, suggesting that a polyoxometalate nanocapsule can form a functional synthetic ion channel in a lipid bilayer membrane.
Self-assembly of cataonic surfactanct DODA around a porous nanocapsule in a hydrophobic solvent (octane). The nanocapsule is shown in red, DODA is shown in blue, the octane solution is not shown. This animation illustrates 164 nanoseconds of molecular dynamics simulation performed using a custom coarse-grained model, which correspond to approximately 10 microseconds in experimental system.
Self-assembly of a liposome-coated metal nanoparticle. A mixture of cationic surfactant (DODA), phospholipid (POPC), water and a porous nanocapsule is simulated using coarsed-grained molecular dynamics (CGMD). The nanocapsule is shown in red, DODA in blue, POPC in white, water and ions are not shown. The animation illustrates 100 nanoseconds of the CGMD trajectory.
The same simulation as above. Select lipid and surfactant molecules are removed to shown the inner structure of the liposome-coated metal nanoparticle. The animation shows the self-assembly process in the reference frame of the metal nanoparticle, which is why the nanoparticle appears to be stationary.
Fusion of a liposome-coated nanoparticle with a lipid bilayer membrane. The cationic surfactant (DODA) and phospholipid (POPC) forming the liposome-coated nanoparticle are shown in blue and white, respectively. The lipid molecules comprising the upper and bottom leaflets of the bilayer at the beginning of the simulation are shown in green and red, respectively. Water and ions are not shown. During the course of the simulation, randomly selected lipid molecules are removed from the top leaflet and inserted into the bottom leaflet to flatten the membrane. This animation illustrates 940 ns of CGMD, which corresponds to approximatley 70 microseconds of experimental time.
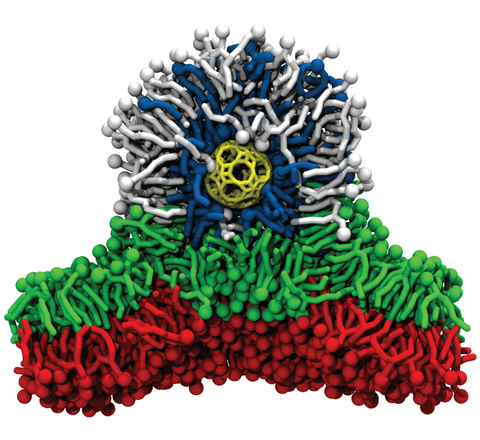
The same simulation as above. Select lipid and surfactant molecules are removed to shown the inner structure of the liposome-coated metal nanoparticle. The porous metal-oxide core particle is shown as yellow wirefram.
A porous metal nanoparticle acts as a membrane-embedded ion channel. The nanoparticle is shown in yellow, cation surfactant (DODA) in dark gray, phospholipid (POPC) bilayer in green, Na+ and Cl− ions in blue and red, respectively, water molecules are shown as white spheres. The animation provides a cut-away view of a 32 ns fragment of the 351 ns CGMD trajectory.