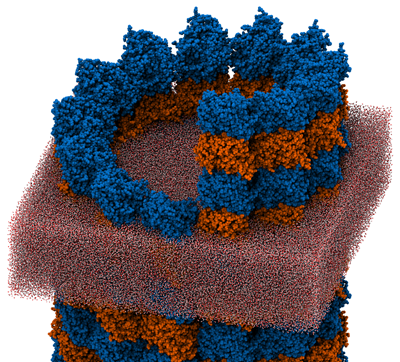
Microtubules (MTs) are the largest type of cellular filament, essential in processes ranging from mitosis and meiosis to flagellar motility. Many of the processes depend critically on the mechanical properties of the MT, but the elastic moduli, notably the Young's modulus, are not directly revealed in experiment, which instead measures either flexural rigidity or response to radial deformation. Molecular dynamics (MD) is a method that allows the mechanical properties of single biomolecules to be investigated through computation. Typically, MD requires an atomic resolution structure of the molecule, which is unavailable for many systems, including MTs. By combining structural information from cryo-electron microscopy and electron crystallography, we have constructed an all-atom model of a complete MT and used MD to determine its mechanical properties. The simulations revealed nonlinear axial stress-strain behavior featuring a pronounced softening under extension, a possible plastic deformation transition under radial compression, and a distinct asymmetry in response to the two senses of twist. This work demonstrates the possibility of combining different levels of structural information to produce all-atom models suitable for quantitative MD simulations, which extends the range of systems amenable to the MD method and should enable exciting advances in our microscopic knowledge of biology.