Single-molecule biophysics experiments in silico: Toward a physical model of a replisome
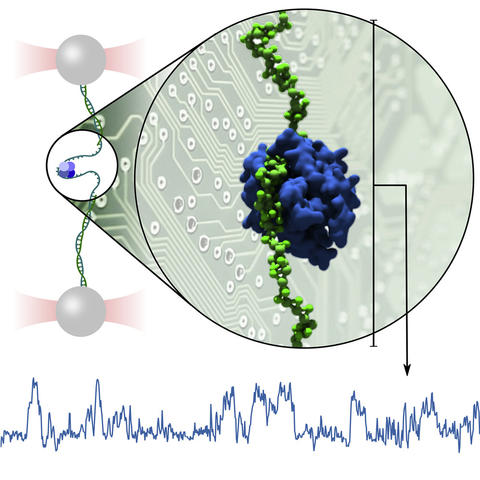
The interpretation of single-molecule experiments is frequently aided by computational modeling of biomolecular dynamics. The growth of computing power and ongoing validation of computational models suggest that it soon may be possible to replace some experiments outright with computational mimics. Here, we offer a blueprint for performing single-molecule studies in silico using a DNA-binding protein as a test bed. We demonstrate how atomistic simulations, typically limited to sub-millisecond durations and zeptoliter volumes, can guide development of a coarse-grained model for use in simulations that mimic single-molecule experiments. We apply the model to recapitulate, in silico, force-extension characterization of protein binding to single-stranded DNA and protein and DNA replacement assays, providing a detailed portrait of the underlying mechanics. Finally, we use the model to simulate the trombone loop of a replication fork, a large complex of proteins and DNA.
All-atom MD simulation of an SSB protein surrounded by 31 dT5 ssDNA fragments. The protein (blue) and the DNA backbone (grey) are shown using a van der Waals representation, while the DNA bases (green) are shown as molecular bonds. Solvent is hidden for clarity. DNA fragments from neighboring periodic unit cell images are shown to avoid flickering associated with a periodic boundary crossing. The movie corresponds to a 250 ns MD trajectory.
Comparison of DNA density in all-atom and CG MD. Average density of ssDNA “P” beads constructed from the results of all-atom MD simulations of the SSB protein surrounded 31 dT5 DNA fragments (green semi-transparent surface) and from the results of CG simulations of the same system (purple semi-transparent surface) carried out using the CG potential maps obtained at the end of the IBI optimization procedure. The SSB core particle and the crystallographically-resolved bound DNA (PDB accession code: 1EYG) are shown using the same representations as in Supplementary Video 1. The animation shows a full rotation of the average density superimposed with a molecular image of the crystallographic structure about a symmetry axis of the SSB protein.
DNA model under tension. CG simulation of a 70 nt ssDNA strand (green ball-and-stick representation) under 5 pN tension. The tension is applied in the vertical directions. The movie covers 4000 ns of the CG simulation.
DNA model with SSB under tension. CG simulation of a 70 nt ssDNA strand (green ball-and-stick representation) under 5 pN tension having an SSB molecule (blue molecular surface) initially bound to the DNA. The movie covers 4000 ns of the CG simulation.
Failed DNA transfer attempt. CG simulation of a 70 nt ssDNA strand (blue) attempting and failing to displace a 65 nt ssDNA strand (green) initially bound to the SSB protein (white molecular surface). The movie covers 25,800 ns of the CG simulation, highlighting a failed transfer attempt.
Successful DNA transfer attempt. CG simulation of a 70 nt ssDNA strand (blue) attempting and successfully displacing a 65 nt ssDNA strand (green) initially bound to the SSB protein (white molecular surface). The movie covers 8755 ns of the CG simulation, highlighting a successful transfer.
Exchange of SSB molecules. CG simulation of the exchange of two SSB molecules (red and blue molecular surfaces) bound to the same DNA construct (green ball-and-stick representation) tethered to a surface and featuring a 70-nt ssDNA strand. The transient distortions of the dsDNA structure result from a trajectory smoothing procedure that was applied to aid visualization of the SSB transfer process. The movie covers 2100 ns of the CG simulation
CG simulation of the trombone loop of the T7 replication fork. Semi-transparent molecular surfaces represent the helicase (teal) in complex with the primase (purple) and polymerase (yellow) proteins, which were resolved in the cryo-EM structure of the complex and represented in the CG simulation using grid-based potentials that acted on the ssDNA and SSB proteins. Double-stranded DNA and ssDNA beads bound to the above proteins are shown in the animated by were not represented explicitly in the simulations. The remaining ssDNA (green beads and rods) was coated with multiple SSB proteins (rainbow colored molecular surface). The movie covers the entire 1600 ns CG simulation.
SSB repositioning during CG simulation of the trombone loop of the T7 replication fork. Highlight of the dissociation and rebinding of an SSB protein from the trombone loop occuring after 7 μs of simulation. The movie covers 350 ns of the CG simulation. The DNA and proteins are depicted using the same representations as in Supplementary Video 8.