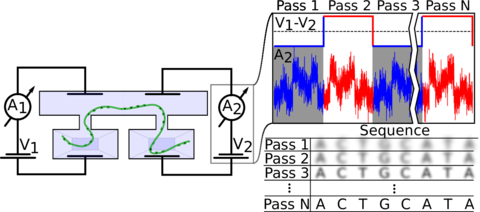
In this article, we describe a proof-of-principle demonstration of a new nanopore sequencing method that directly resolves the shortcomings of the current nanopore sequencing technology. Our method is built around a double-nanopore system that enables infinite-depth sequencing of individual DNA or RNA molecules by moving the same molecules repeatedly back and forth (‘flossing’) through the nanopores, a process that can be repeated as often as needed until the desired precision of nucleotide determination is reached (see SI Movie 1 and 2). In doing so, our approach removes the need for any error-prone enzyme for speed control and finally provides a feasible path for the development of a purely solid-state nanopore sequencing platform, a long-promised game-changer in the nanopore sequencing field. Our double-nanopore sequencing platform features a nanofluidic system for high-fidelity delivery and threading of native nucleic acids, enabling analysis of genomic-length DNA and RNA. The combination of high-fidelity loading and repeat sequencing of the same molecules allows for direct sequencing of DNA and RNA, with order of magnitude improvements in the performance over the current state-of-the-art nanopore sequencing with regard to ion-current signal and base calling at a truly single-molecule level.