Molecular Transport across the Ionic Liquid−Aqueous Electrolyte Interface in a MoS2 Nanopore
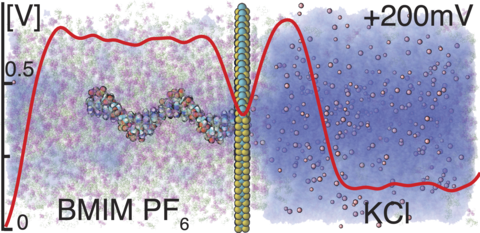
Nanopore sequencing of DNA has been enabled by the use of a biological enzyme to thread DNA through an engineered biological nanopore while recording the ionic current flowing through the nanopore. Efforts to realize a similar concept using a solid-state nanopore have been met with several technical challenges, one of which is the high speed of DNA translocation and the other the low ionic current contrast among individual nucleotides. A promising avenue to addressing both problems is using an ionic liquid to slow DNA translocation and a tiny nanopore in the MoS2 membrane to distinguish individual nucleotides. The physical mechanisms enabling these technical advances have remained elusive. Here, we characterize the ion and DNA transport through the ionic liquid/aqueous electrolyte interface, with and without a MoS2 nanopore, using the all-atom molecular dynamics method. We find that the partial miscibility of the ionic liquid and the aqueous electrolyte considerably alters the physics of the nanopore translocation process. Thus, the interface of the two phases generates a contact potential of 600 mV, the ionic current is dominated by the motion of ionic liquid molecules through the aqueous solution phase, and the DNA nucleotides exhibit preferential partitioning into the aqueous electrolyte, which leads to spontaneous transport of DNA polymers from the ionic liquid to the aqueous solution compartment in the absence of external voltage bias. The complex physics of the two-phase nanopore system offers a multitude of opportunities for extending the functionality of nanopore-sensing platforms.
Supplementary Movie 1. Phase separation of initially homogeneously system containing equal volumes of [Bmim][PF6] and 1M KCl solutions. The movie represents a 160 ns MD trajectory. Spheres illustrate K and Cl ions (pink), atoms of BMIM (green) and PF6 (magenta); water is shown as a blue semi-transparent surface
Supplementary Movie 2. Equilibration of a [Bmim][PF6]/1M KCl system. This equilibrium MD simulation begins with a state where two equal volumes of pure [Bmim][PF6] and 1M KCl electrolyte are in contact. The movie represents a 170 ns MD trajectory. Red and white spheres illustrate oxygen and hydrogen atoms of water, translucent green and gray spheres correspond to atoms of BMIM and PF6; pink spheres illustrate K and Cl ions.
Supplementary Movie 3. MD simulation of the two-phase [Bmim][PF6]/1M KCl system under a 200 mV bias. Atoms of BMIM and PF6 are shown as magenta and green spheres, K and Cl ions in pink, water as a blue semitransparent surface. The simulation begins from a configuration obtained at the end of a 170 ns equilibration (Supplementary Movie 2). The movie illustrates a 120 ns MD trajectory.
Supplementary Movie 4. Ion transport through water networks in the ionic liquid-rich phase. The simulation illustrates the same trajectory as the Supplementary Movie 3, where a 200 mV voltage was applied across the system normal to the ionic liquid/aqueous solution interface. In this movie, water is represented as red translucent spheres, K and Cl ions as gold and cyan spheres, respectively, whereas BMIM and PF6 are not shown for clarity.
Supplementary Movie 5. Reversible voltage-dependent switching of the interface geometry. Green and blue vdW spheres represent K and Cl ions, respectively, gray represents BMIM and PF6, water is not shown for clarity. The simulation begins with a two-phase conformation obtained at the end of a 120 ns simulations under a 200 mV bias. A transmembrane bias of 1.0 V is applied for 76 ns. Following that, the system is equilibrated in the absence of external electric field for 100 ns.
Supplementary Movie 6. Single nucleotide dynamics in [Bmim][PF6]. Twenty adenine, cytosine, guanine and thymine nucleotides were randomly placed in pure [Bmim][PF6] and equilibrated for 260 ns. Each nucleotide is shown in a different color, the atoms of BMIM and PF6 as translucent magenta and green spheres, respectively.
If the problem persists contact your website administrator. Please check logs for further debugging.
Supplementary Movie 7. Single nucleotide dynamics in KCl solution. Twenty adenine, cytosine, guanine and thymine nucleotides were randomly placed in 1M KCl electrolyte and equilibrated for 100 ns. Each nucleotide is shown in a different color, water as a semitransparent surface. K and Cl ions are not shown for clarity.
Supplementary Movie 8. Equilibration of DNA homopolymers in pure [Bmim][PF6]. The initial conformation of each homopolymer was taken from a B-form DNA duplex. Each DNA base is uniquely colored and atoms are represented by spheres. BMIM and PF6 molecules are represented by magenta and green translucent spheres, respectively.
Supplementary Movie 9. Electrolyte-driven aggregation of four hydrated single adenine nucleotides in ionic liquid. Four adenine nucleotides (red spheres) are placed randomly, separated by at least 3 nm. Nucleotides were initially hydrated by a 1.5 nm sphere of water molecules represented as red (oxygen) and white (hydrogen) spheres. The movie illustrates four 150 ns MD trajectories differing by KCl concentration.
Supplementary Movie 10. MD simulation of three adenine nucleotides in a two-phase system under a 200 mV transmembrane bias. The two-phase system was equilibrated for 100 ns before the nucleotides were insterted at random position. Following that, the system was equilibrated for additional 30 ns. The nucleotides are represented as cyan spheres whereas BMIM and PF6 are are shown as semitransparent molecules colored in green and magenta. Water and KCl are not shown for clarity. The movie represents a 80 ns MD trajectory.
Supplementary Movie 11. Spontaneous transport of ssDNA homopolymers through a MoS2 nanopore. In each simulation system, a poly(dA)20, poly(dC)20, poly(dG)20 or poly(dT)20 strand was inserted into a MoS2 nanopore system, which was then equilibrated for 15 ns with all non-hydrogen atoms of the DNA constrained to their initial positions. The DNA is shown us- ing spheres colored according to the atom type: blue (nitrogen), red (oxygen), white (hydrogen), carbon (cyan) and gold (phosphorous). This movie represents 190 ns of simulation time.