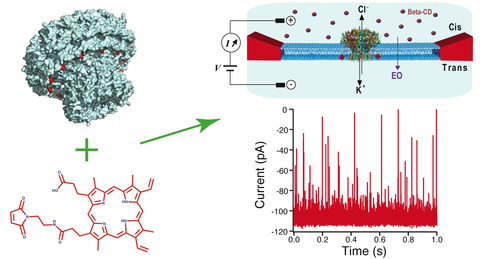
Nanopore-based sensors for nucleic acid sequencing and single-molecule detection typically employ pore-forming membrane proteins with hydrophobic external surfaces, suitable for insertion into a lipid bilayer. In contrast, hydrophilic pore-containing molecules such as DNA origami, have been shown to require chemical modification to favor insertion into a lipid environment. In this work, we describe a strategy for inserting polar proteins with an inner pore into lipid membranes, focusing here on a circular 12-subunit assembly of the thermophage G20c portal protein. X-ray crystallography, electron microscopy, molecular dynamics and thermal/chaotrope denaturation experiments all find the G20c portal protein to have a highly stable structure, favorable for nanopore sensing applications. Porphyrin conjugation to a cysteine mutant in the protein facilitates the protein’s insertion into lipid bilayers, allowing us to probe ion transport through the pore. Finally, we probed the portal interior size and shape using a series of cyclodextrins of varying sizes, revealing asymmetric transport that possibly originates from the portal’s DNA-ratchet function.