Nanopore Sensing of Protein Folding
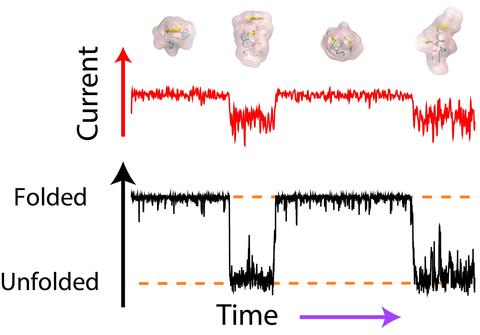
Single-molecule studies of protein folding hold keys to unveiling protein folding pathways and elusive intermediate folding states-attractive pharmaceutical targets. Although conventional single-molecule approaches can detect folding intermediates, they presently lack throughput and require elaborate labeling. Here, we theoretically show that measurements of ionic current through a nanopore containing a protein can report on the protein’s folding state. Our all-atom molecular dynamics (MD) simulations show that the unfolding of a protein lowers the nanopore ionic current, an effect that originates from the reduction of ion mobility in proximity to a protein. Using a theoretical model, we show that the average change in ionic current produced by a folding−unfolding transition is detectable despite the orientational and conformational heterogeneity of the folded and unfolded states. By analyzing millisecond-long all-atom MD simulations of multiple protein transitions, we show that a nanopore ionic current recording can detect folding−unfolding transitions in real time and report on the structure of folding intermediates.
This animation illustrates a 0.2 millisecond all-atom molecular dynamics simulation of a Trp-cage protein undergoing folding-unfolding transitions inside a solid-state nanopore. The simulation begins having the protein in a folded state. The two traces on the right show the simulated ionic current flowing through the nanopore and the fraction of native contacts (Q) in the protein conformation as a function of time. The Q values for the folded and unfolded states are 1 and 0, respectively. The ionic current calculations were performed for a 300 mV transmembrane bias and a 2 M KCl solution. The protein is seen to block the ionic current more when it adopts an unfolded conformation.