Molecular dynamics study of MspA arginine mutants predicts slow DNA translocations and ion current blockades indicative of DNA sequence
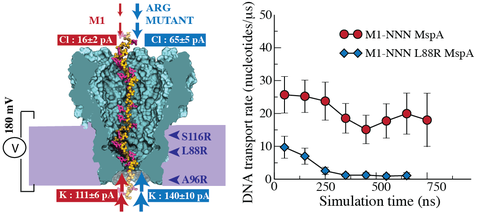
The protein nanopore Mycobacteria smegmatis porin A (MspA), can be used to sense individual nucleotides within DNA, potentially enabling a technique known as nanopore sequencing. In this technique, single-stranded DNA electrophoretically moves through the nanopore and results in an ionic current that is nucleotide-specific. However, with a high transport velocity of the DNA within the nanopore, the ionic current cannot be used to distinguish signals within noise. Through extensive (~100 μs in total) all-atom molecular dynamics simulations, we examine the effect of positively charged residues on DNA translocation rate and the ionic current blockades in MspA. Simulation of several arginine mutations show a ~10-30 fold reduction of DNA translocation speed without eliminating the nucleotide induced current blockages. Comparison of our results with similar engineering efforts on a different nanopore (α-hemolysin) reveals a nontrivial effect of nanopore geometry on the ionic current blockades in mutant nanopores.
A molecular dynamics trajectory of a poly(dC) DNA strand permeation through a truncated M1-NNN pore driven by a 180 mV transmembrane bias. About 30 nucleotides are seen to permeate through the MspA constriction in less than 800 ns.
A 4 μs molecular dynamics trajectory of a poly(dC) strand permeating through a truncated L88R/T83R/S116R mutant pore at a 180 mV transmembrane bias. The permeation of the strand appears to be halted owing to the formation of contacts with the arginine residues on the wall of the pore (shown in blue).