Ion Channels Made from a Single Membrane-Spanning DNA Duplex
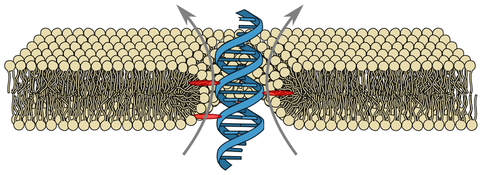
Due to their hollow interior, transmembrane channels are capable of opening up pathways for ions across lipid membranes of living cells. Here, we demonstrate ion conduction induced by a single DNA duplex that lacks a hollow central channel. Decorated with six porpyrin-tags, our duplex is designed to span lipid membranes. Combining electrophysiology measurements with all-atom molecular dynamics simulations, we elucidate the microscopic conductance pathway (see water channel trajectory, require login to nanoHUB). Ions flow (see ion transport trajectory) at the DNA-lipid interface as the lipid head groups tilt towards the amphiphilic duplex forming a toroidal pore filled with water and ions. Ionic current traces produced by the DNA-lipid channel show well-defined insertion steps, closures and gating similar to those observed for traditional protein channels or synthetic pores. Ionic conductances obtained through simulations and experiments are in excellent quantitative agreement. The conductance mechanism realized here, with the smallest possible DNA-based ion channel, offers a route to design a new class of synthetic ion channels with maximum simplicity.
Molecular dynamics simulation of the DNA duplex in a lipid bilayer membrane. The DNA strands are coloured in blue and yellow; the porphyrin groups are coloured in grey. The lipid molecules are drawn as lines; C, N, O and P atoms are coloured in green, blue, red and ochre, respectively. The hydrogen atoms of lipid molecules are not shown. For clarity, lipid molecules facing the viewer are removed. The slide bar at the upper right corner indicates the progression of the trajectory in nanosecond (ns). The first 200 ns of the simulation correspond to equilibration (no applied bias); the remaining part of the movie illustrates a trajectory at +100 mV bias. At the beginning of the movie, a static image illustrates the initial placement of the DNA duplex within the lipid membrane, highlighting the location of the porphyrin groups. The second stage of the movie illustrates the stochastic displacements of waters molecules around the duplex; O and H atoms of water are shown as red and white spheres, respectively. The final part of the movie illustrates the transmembrane transports. All lipid molecules are hidden; the volume occupied by water is represented by a semi-transparent surface; potassium ions are shown as green spheres. For clarity, only those potassium ions that showed considerable displacement across the lipid bilayer (top 10%) are shown.