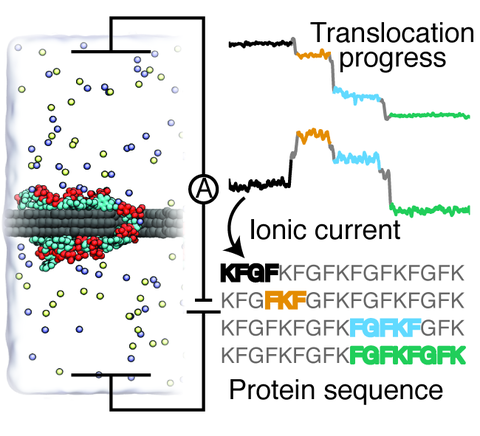
An inexpensive, reliable method for protein sequencing is essential to unraveling the biological mechanisms governing cellular behavior and disease. Current protein sequencing methods suffer from limitations associated with the size of proteins that can be sequenced, the time, and the cost of the sequencing procedures. This study reports the results of all-atom molecular dynamics simulations that investigated the feasibility of using graphene nanopores for protein sequencing. The study is focused on the biologically significant phenylalanine-glycine repeat peptides (FG-nups)—parts of the nuclear pore transport machinery. Surprisingly, FG-nups are found to behave similarly to single stranded DNA: The peptides adhere to graphene and exhibit stepwise translocation when subject to a transmembrane bias or a hydrostatic pressure gradient. Reducing the peptide's charge density or increasing the peptide's hydrophobicity is found to decrease the translocation speed. Yet, unidirectional and stepwise translocation driven by a transmembrane bias is observed even when the ratio of charged to hydrophobic amino acids is as low as 1:8. The nanopore transport of the peptides is found to produce stepwise modulations of the nanopore ionic current correlated with the type of amino acids present in the nanopore, suggesting that protein sequencing by measuring ionic current blockades may be possible.