The structure and intermolecular forces of DNA condensates
Spontaneous assembly of DNA molecules into compact structures is ubiquitous in biological systems. Experiment has shown that polycations can turn electrostatic self-repulsion of DNA into attraction, yet the physical mechanism of DNA condensation has remained elusive. Here, we report the results of atomistic molecular dynamics simulations that elucidated the microscopic structure of dense DNA assemblies and the physics of interactions that makes such assemblies possible. Reproducing the setup of the DNA condensation experiments, we measured the internal pressure of DNA arrays as a function of the DNA–DNA distance, showing a quantitative agreement between the results of our simulations and the experimental data. Analysis of the MD trajectories determined the DNA–DNA force in a DNA condensate to be pairwise, the DNA condensation to be driven by electrostatics of polycations and not hydration, and the concentration of bridging cations, not adsorbed cations, to determine the magnitude and the sign of the DNA–DNA force. Finally, our simulations quantitatively characterized the orientational correlations of DNA in DNA arrays as well as diffusive motion of DNA and cations.
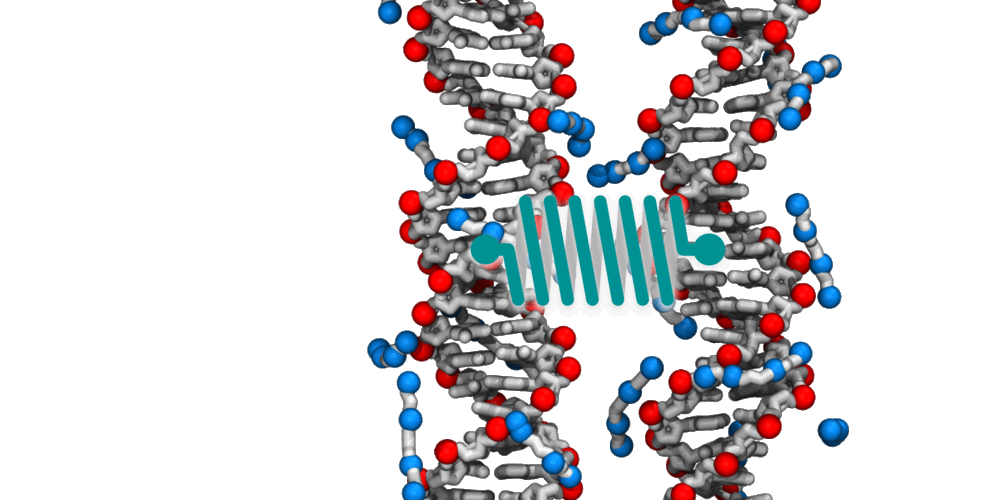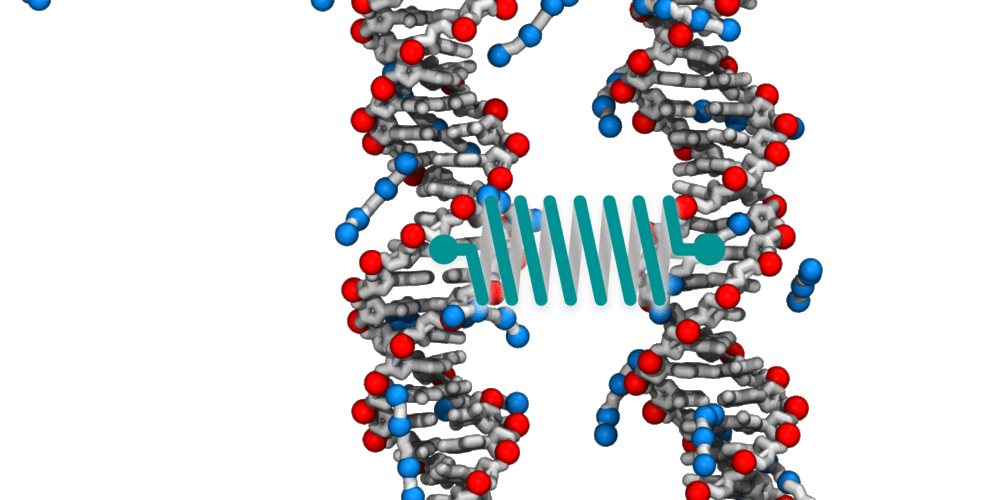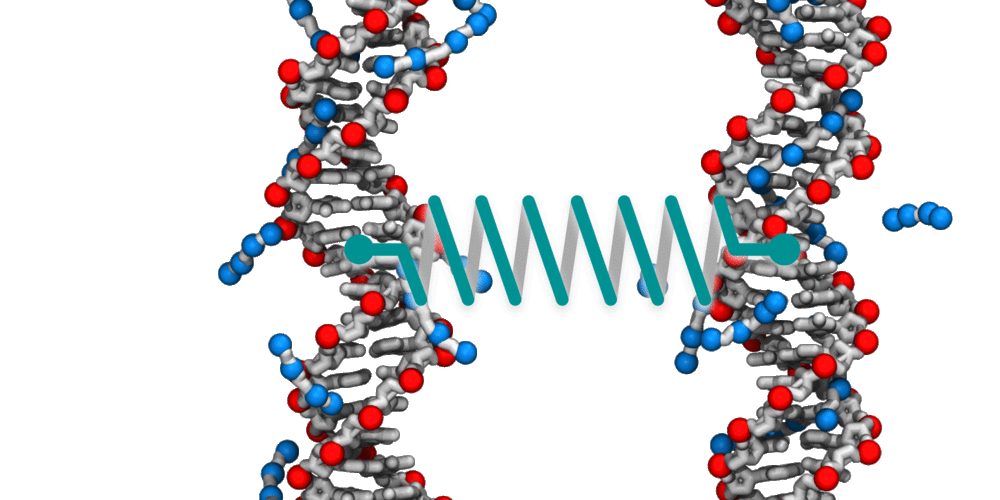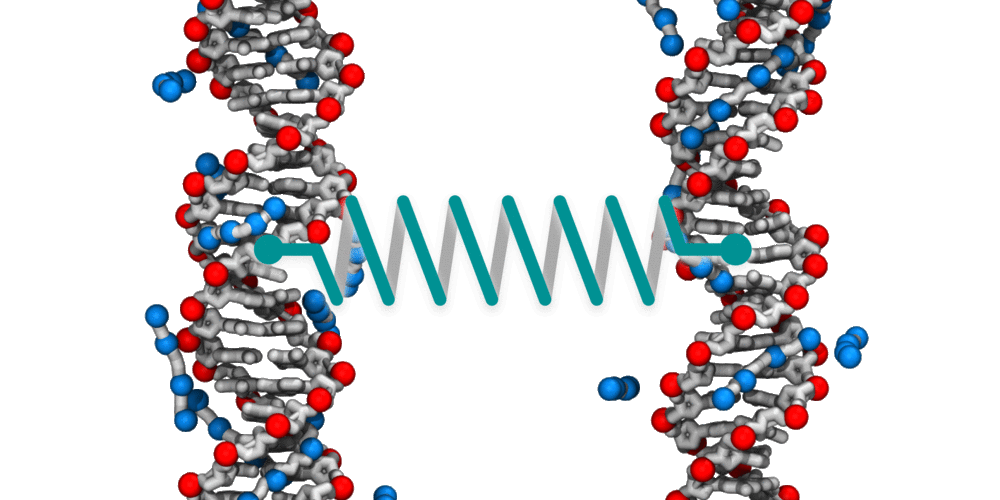
Animation illustrating the use of molecular dynamics simulation to evaluate effective force between two DNA molecules. A harmonic potential (illustrated by a spring) is applied between the centers of mass of the two DNA molecules. The average deviation of the center of mass distance from its equilibrium value reports the effective force. The DNA molecules are free to rotate and translate with respect to one another. The four blue spheres connected by gray bonds represent a molecules of spermine, a +4 cation.
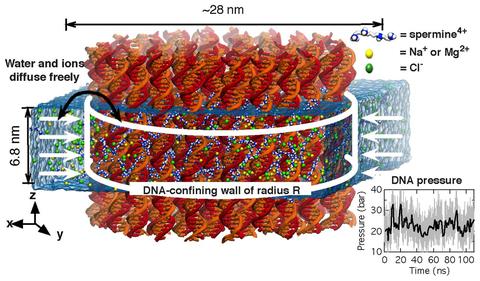
Molecular dynamics simulation of a DNA array system. A system of 64 DNA helices is placed in electrolyte solution containing spermine molecules (blue and gray) and background electrolyte (NaCl, orange and yellow). The DNA molecules are confined to remain within a cylinder of a prescribed radius, water and ions can move in and out of the array. The average total force exerted by the confining potential on the DNA reports on the internal pressure of the array. At the beginning of the simulation, all spermine molecules are located outside of the DNA array. The movie illustrates a 200 ns molecular dynamics trajectory.