Molecular Dynamics of Membrane-Spanning DNA Channels: Conductance Mechanism, Electro-Osmotic Transport, and Mechanical Gating
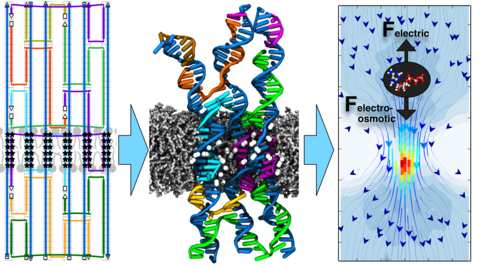
DNA self-assembly has emerged as a new paradigm for design of biomimetic membrane channels. Several experimental groups have already demonstrated assembly and insertion of DNA channels into lipid bilayer membranes; however, the structure of the channels and their conductance mechanism have remained undetermined. Here, we report the results of molecular dynamics simulations that characterized the biophysical properties of the DNA membrane channels with atomic precision. We show that, while overall remaining stable, the local structure of the channels undergoes considerable fluctuations, departing from the idealized design. The transmembrane ionic current flows both through the central pore of the channel as well as along the DNA walls and through the gaps in the DNA structure. Surprisingly, we find that the conductance of DNA channels depend on the membrane tension, making them potentially suitable for force-sensing applications. Finally, we show that electro-osmosis governs the transport of druglike molecules through the DNA channels.
Movie 1. Internal structure of the DNA origami channel (System I) at the end of the 70-ns equilibration simulation. The DNA backbone and bases are shown in green and blue, respec- tively. Ethyl-modifications are shown with a vdW representation (carbon, cyan; hydrogen, white). Lipid molecules are shown with white molecular bonds.
Movie 2. Structural fluctuation of the DNA channel in System I during the last 13 ns of the 40 ns production simulation carried out in the absence of applied electric field or mechanical stress. Each DNA strand has a unique color. Lipid molecules are shown using a molecular bond representation (carbon, cyan; oxygen, red; nitrogen, blue; phosphorus, ochre).
Movie 3. Transmembrane transports of K+ (blue spheres) and Cl− (red spheres) ions through the pore of the DNA origami channel (System I) at a 600 mV transmembrane bias; the movie covers a 5 ns fragment of the MD trajectory. This simulation was carried out under the zero tension lipid bilayer condition. DNA is shown in gray, lipid and water molecules are not shown for clarity.
Movie 4. A typical trajectory of an ATP molecule that passes from one size of the membrane to the other through the transmembrane pore in System I. The ATP molecule is seen to move in the direction of the applied electric field (opposite to the direction prescribed by its negative charge). The movie covers a 300 ns MD trajectory. DNA backbone and bases are shown in green cartoon and blue molecular-bonds representations, respectively. An ATP molecule is shown in spheres colored by atom types: nitrogen, blue; oxygen, red; carbon, cyan; hydrogen, white. Lipid molecules are shown in gray molecular bonds.