Rapid and Accurate Determination of Nanopore Ionic Current Using a Steric Exclusion Model
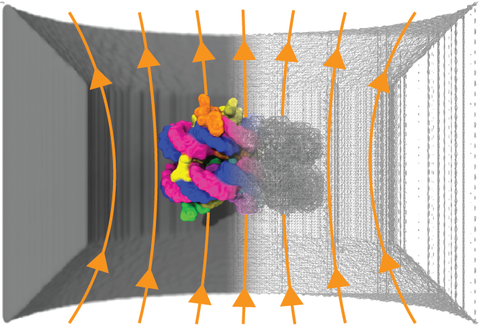
Nanopore sensing has emerged as a versatile approach to detection and identification of biomolecules. Presently, researchers rely on experience and intuition for choosing or modifying the nanopores to detect a target analyte. The field would greatly benefit from a computational method that could relate the atomic-scale geometry of the nanopores and analytes to the blockade nanopore currents they produce. Existing computational methods are either computationally too expensive to be used routinely in experimental laboratories or not sensitive enough to account for the atomic structure of the pore and the analytes. Here, we demonstrate a robust and inexpensive computational approach—the steric exclusion model (SEM) of nanopore conductance—that is orders of magnitude more efficient than all-atom MD and yet is sensitive enough to account for the atomic structure of the nanopore and the analyte. The method combines the computational efficiency of a finite element solver with the atomic precision of a nanopore conductance map to yield unprecedented speed and accuracy of ionic current prediction. We validate our SEM approach through comparison with the current blockades computed using the all-atom molecular dynamics method for a range of proteins confined to a solid-state nanopore, biological channels embedded in a lipid bilayer membranes, and blockade currents produced by DNA homopolymers in MspA. We illustrate potential applications of SEM by computing blockade currents produced by nucleosome proteins in a solid-state nanopore, individual amino acids in MspA, and by testing the effect of point mutations on amino acid distinguishability. We expect our SEM approach to become an integral part of future development of the nanopore sensing field.