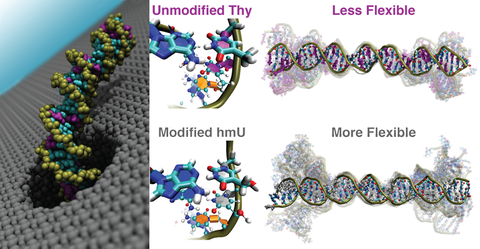
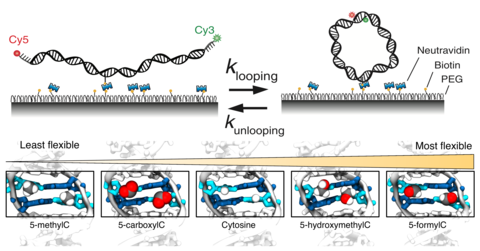
Effect of DNA modifications on the flexibility of double-stranded DNA
All eukaryotes use DNA to store their genetic information, and the nucleotide sequence of DNA encodes for all possible functions of a biological cell. Beyond the mere nucleotide sequence, it is the set of epigenetic markers that determines which parts of the genetic code are active, ultimately controlling the cell’s fate. The most common epigenetic marker is methylation of DNA; however, other DNA modifications have been discovered that may play a role in the development and differentiation of cells. Methods for reliably detecting these modifications are crucial to furthering our understanding of the epigenetic processes that are undergone in cells. One physical property of DNA that changes when DNA modifications are present is flexibility, and changes in local flexibility could directly modulate gene activity. Here we use two single-molecule techniques along with molecular dynamics to study how DNA flexibility is affected by modifications to the DNA bases.
The nanopore method measures the ability of double-stranded DNA to pass through a small hole in a membrane that has about the same diameter as the diameter of the DNA itself. DNA that is more flexible, is able to navigate through a nanopore faster than DNA that is more rigid. Our experimental collaborators in the Wanunu group measured the transit times of pieces of DNA electrophoretically driven through a nanometer-sized hole in a membrane. They found that modified DNA (Changing all Thymine to hydroxymethylUracil) passes through the nanopore faster under the same bias.
In the DNA cyclization assay, a double-stranded DNA molecule was constructed so that it could form a loop. Our experimental colleagues in the Ha group were able to measure the time that it took this DNA to form a loop. The faster the loop formed, the more flexible the DNA molecule was expected to be. Using carefully constructed DNA with modifications to the cytosine bases, the effect of the DNA modifications could be ranked, with some modifications stiffening the DNA, and others making the DNA more flexible.
We use all-atom molecular dynamics simulations to characterize the differences in flexibility of DNA with and without one of several DNA modifications. To do this, we create an all-atom model of a fragment of DNA submerged in an ionic solution similar to that used in experiments. We simulate the DNA without restraints and analyze the fluctuations that the DNA undergoes during the simulation using x3DNA analysis. By performing two simulations with identical DNA except for a modification, we can directly compare the effect that the modification has on the flexibility of the DNA.