Multi-resolution simulation of DNA transport through large synthetic nanostructures.
Modeling and simulation has become an invaluable partner in development of nanopore sensing systems. The key advantage of the nanopore sensing method - the ability to rapidly detect individual biomolecules as a transient reduction of the ionic current flowing through the nanopore - is also its key deficiency, as the current signal itself rarely provides direct information about the chemical structure of the biomolecule. Complementing experimental calibration of the nanopore sensor readout, coarse-grained and all-atom molecular dynamics simulations have been used extensively to characterize the nanopore translocation process and to connect the microscopic events taking place inside the nanopore to the experimentally measured ionic current blockades. Traditional coarse-grained simulations, however, lack the precision needed to predict ionic current blockades with atomic resolution whereas traditional all-atom simulations are limited by the length and time scales amenable to the method. Here, we describe a multi-resolution framework for modeling electric field-driven passage of DNA molecules and nanostructures through to-scale models of synthetic nanopore systems. We illustrate the method by simulating translocation of double-stranded DNA through a solid-state nanopore and a micron-scale slit, capture and translocation of single-stranded DNA in a double nanopore system, and modeling ionic current readout from a DNA origami nanostructure passage through a nanocapillary. We expect our multi-resolution simulation framework to aid development of the nanopore field by providing accurate, to-scale modeling capability to research laboratories that do not have access to leadership supercomputer facilities.
Coarse-grained molecular dynamics simulation of double-stranded DNA passage through a solid-state nanopore (goes with Figure 1 of the publication)
Coarse-grained simulation of double-stranded DNA passage through a graphene slit (goes with Figure 2 of the publication)
Coarse-grained molecular dynamics simulation of single-stranded DNA trapping in a dual nanopore systems (goes with Figure 3 of the publication)
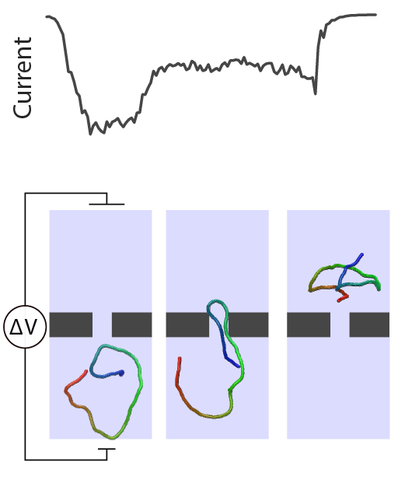
Coarse-grained simulation of DNA construct's capture and expulsion by a nanocapillary. The DNA construct consists of a nicked double-stranded DNA decorated with double-stranded overhangs. This movie illustrates the process considered in Figure 4 of the publication.
Coarse-grained simulation of a DNA origami plate transport through a nanocapillary. This movie illustrates the process considered in Figure 5 of the publication.