Discrimination of RNA fiber structures using solid-state nanopores
RNA fibers are a class of biomaterials that can be assembled using HIV-like kissing loop interactions. Because of the programmability of molecular design and low immunorecognition, these structures present an interesting opportunity to solve problems in nanobiotechnology and synthetic biology. However, the experimental tools to fully characterize and discriminate among different fiber structures in solution are limited. Herein, we utilize solid-state nanopore experiments and Brownian dynamics simulations to characterize and distinguish several RNA fiber structures that differ in their degrees of branching. We found that, regardless of the electrolyte type and concentration, fiber structures that have more branches produce longer and deeper ionic current blockades in comparison to the unbranched fibers. Experiments carried out at temperatures ranging from 20–60 °C revealed almost identical distributions of current blockade amplitudes, suggesting that the kissing loop interactions in fibers are resistant to heating within this range.
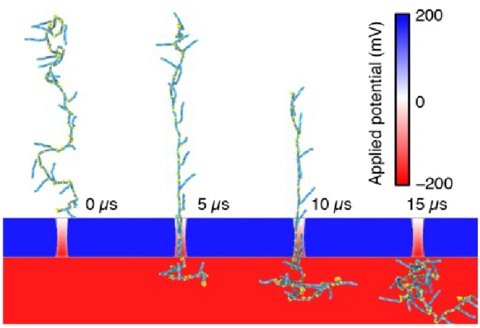
Coarse-grained Brownian dynamics simulation of a non-functionalized (NF) RNA fiber passing through a 4.5 nm diameter nanopore. COMSOL Multiphysics was used to determine the distribution of the electrostatic potential in and around the nanopore. The membrane containing a nanopore was represented as a steric (repulsive) potential.
Coarse-grained Brownian dynamics simulation of a fiber with branching resulting from functionalization with dicer substrate RNAs either at every other monomer (EOM) passing through a 4.5 nm diameter nanopore.
Coarse-grained Brownian dynamics simulation of a fiber with branching resulting from functionalization with dicer substrate RNAs either at every monomer (EM) passing through a 4.5 nm diameter nanopore.
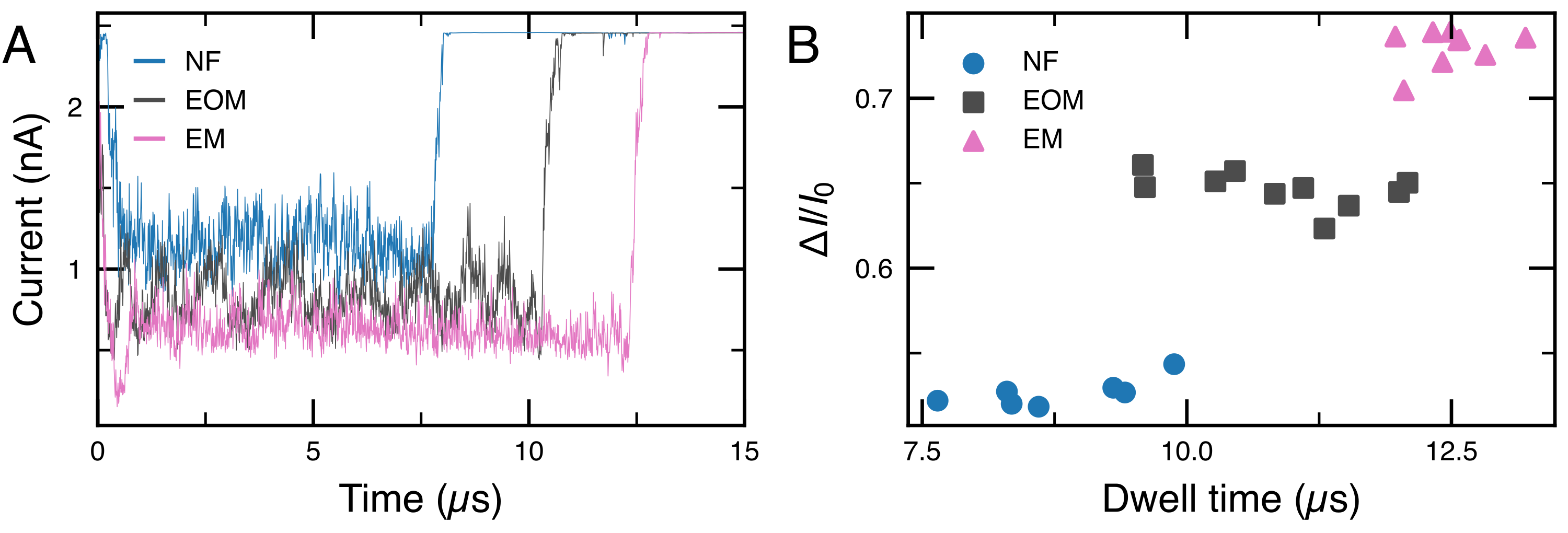
Ionic current blockade from coarse-grained Brownian dynamics simulation of RNA fibers passing through a 4.5 nm diameter nanopore. (A) Typical ionic current traces calculated from the translocation trajectories using SEM23,24. (B) Average relative blockade current amplitude versus dwell time for each translocation trajectory. The different fiber types are readily distinguished.