Hydrophobic Interactions between DNA Duplexes and Synthetic and Biological Membranes.
Equipping DNA with hydrophobic anchors enables targeted interaction with lipid bilayers for applications in biophysics, cell biology, and synthetic biology. Understanding DNA-membrane interactions is crucial for rationally designing functional DNA. Here we study the interactions of hydrophobically tagged DNA with synthetic and cell membranes using a combination of experiments and atomistic molecular dynamics (MD) simulations. The DNA duplexes are rendered hydrophobic by conjugation to a terminal cholesterol anchor or by chemical synthesis of a charge-neutralized alkyl-phosphorothioate (PPT) belt. Cholesterol-DNA tethers to lipid vesicles of different lipid compositions and charges, while PPT DNA binding strongly depends on alkyl length, belt position, and headgroup charge. Divalent cations in the buffer can also influence binding. Our MD simulations directly reveal the complex structure and energetics of PPT DNA within a lipid membrane, demonstrating that longer alkyl-PPT chains provide the most stable membrane anchoring but may disrupt DNA base paring in solution. When tested on cells, cholesterol-DNA is homogeneously distributed on the cell surface, while alkyl-PPT DNA accumulates in clustered structures on the plasma membrane. DNA tethered to the outside of the cell membrane is distinguished from DNA spanning the membrane by nuclease and sphingomyelinase digestion assays. The gained fundamental insight on DNA-bilayer interactions will guide the rational design of membrane-targeting nanostructures.
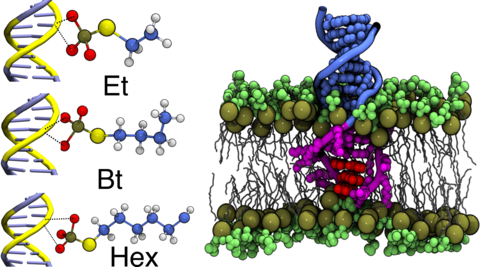
All-atom molecular dynamics simulation of ethyl, butyl and hexyl modified dsDNA molecules centrally anchored to a POPC lipid bilayer membrane. The movie shows a cut-away view of 1 μs long equilibration trajectories for three separate systems. The unmodified and unmodified DNA nucleotides are shown in blue and magenta respectively. The DNA backbone is shown as thin tubes whereas nucleotide base atoms are represented using vdW spheres. Non-hydrogen atoms of the amine-phosphate lipid headgroups are shown as green spheres whereas the lipid tails are shown as white lines. Water and ions are not shown for clarity
All-atom molecular dynamics simulation of terminally modified ethyl, butyl and hexyl dsDNA molecules anchored into the POPC lipid bilayer membrane. The movie shows a cut- away view of 1 μs long equilibration trajectories for three separate systems. The unmodified and unmodified DNA nucleotides are shown in blue and magenta respectively. The DNA backbone is shown as thin tubes whereas nucleotide base atoms are represented using vdW spheres. Non-hydrogen atoms of the amine-phosphate lipid headgroups are shown as green spheres whereas the lipid tails are shown as white lines. Water and ions are not shown for clarity.
All-atom MD simulation of unmodified DNA initially anchored in a POPC lipid bilayer membrane. The DNA is seen to escape from the membrane in less than 400 ns, the simulation duration. The DNA backbone is shown as thin blue tubes whereas nucleotide base atoms are represented using vdW spheres. Non-hydrogen atoms of the amine-phosphate lipid headgroups are shown as green spheres whereas the lipid tails are shown as white lines. Water and ions are not shown for clarity.
SMD pulling of terminally anchored DNA constructs from a lipid bilayer membrane. The movie illustrates cut-away view of four separate SMD simulations each being 100 ns long. The unmodified and unmodified DNA nucleotides are shown in blue and yellow, respectively. The DNA backbone is shown as thin tubes whereas nucleotide base atoms are represented using vdW spheres. Non-hydrogen atoms of the amine-phosphate lipid headgroups are shown as green spheres whereas the lipid tails are shown as white lines. Water and ions are not shown for clarity.