Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore
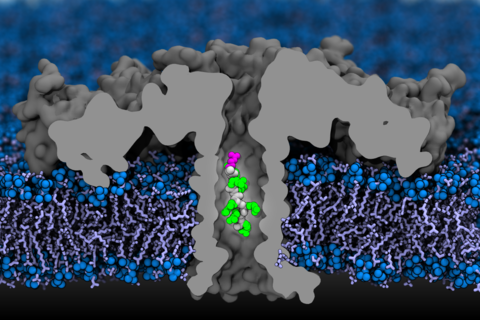
Efforts to sequence single protein molecules in nanopores have been hampered by the lack of techniques with sufficient sensitivity to discern the subtle molecular differences among all twenty amino acids. Here we report ionic current detection of all twenty proteinogenic amino acids in an aerolysin nanopore with the help of a short polycationic carrier. Application of molecular dynamics simulations revealed that the aerolysin nanopore has a built-in single-molecule trap that fully confines a polycationic carrier-bound amino acid inside the sensing region of the aerolysin. This structural feature means that each amino acid spends sufficient time in the pore for sensitive measurement of the excluded volume of the amino acid. We show that distinct current blockades in wild-type aerolysin can be used to identify 13 of the 20 natural amino acids. Furthermore, we show that chemical modifications, instrumentation advances and nanopore engineering offer a route toward identification of the remaining seven amino acids. These findings may pave the way to nanopore protein sequencing.
Research Highlights
1. Tang, Lei. "Sensing proteinogenic amino acids." Nature Methods 17(2):126 (2020) DOI: 10.1038/s41592-020-0741-z
2. Howorka, Stefan, and Zuzanna S. Siwy. "Reading amino acids in a nanopore." Nature Biotechnology 38(2):159 (2020) DOI: 10.1038/s41587-019-0401-y
3. Nanopores can identify the amino acids in proteins, the first step to sequencing, Highlight by Illinois news bureau.
Equilibration of aerolysin nanopore in DPhPC membrane. The video illustrates a 10 ns MD trajectory of the aerolysin system consisting of the aerolysin nanopore (grey, cut-away molecular surface) embedded in a DPhPC lipid bilayer (red for head groups and cyan for tails) and 1 M KCl solution (not shown for clarity).
MD simulation of open pore ionic current. The video illustrates a 2 ns fragment of an MD trajectory of the aerolysin system under a transmembrane bias of −100 mV. The aerolysin nanopore is shown using a grey, cut-away molecular surface; the head groups and tails of the DPhPC lipid bilayer are shown in red and cyan, respectively; the volume occupied by 1 M KCl electrolyte is represented by a blue translucent surface; potassium and chloride ions are shown as violet and green spheres, respectively.
SMD simulation of RR7 peptide translocation through aerolysin. The video illustrates a 100 ns MD trajectory where an RR7 peptide was driven through the aerolysin nanopore using the SMD protocol. The aerolysin nanopore is shown as a grey, cutaway molecular surface; the DPhPC lipid bilayer is shown in red (head groups) and cyan (lipid tails); the backbone of the RR7 peptide is shown in grey while the side chains of all arginine residues are shown in blue except for the last one, which is shown in red.