Electro-Mechanical Conductance Modulation of a Nanopore Using a Removable Gate
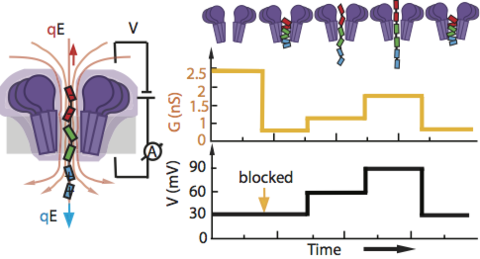
Ion channels form the basis of information processing in livings cells by facilitating exchange of electrical signals across and along cellular membranes. Applying the same principles to man-made systems requires development of synthetic ion channels that can alter their conductance in response to a variety of external manipulations. By combining single-molecule electrical recordings with all-atom molecular dynamics simulations, we here demonstrate a new hybrid nanopore system that allows for both a stepwise change of its conductance and a non-linear current-voltage dependence. The conductance modulation is realized by using a short flexible peptide gate that carries opposite electric charge at its ends. We show that a constant transmembrane bias can position, and in a later stage remove, the peptide gate right at the most sensitive sensing region of a biological nanopore FraC, thus partially blocking its channel and producing a stepwise change in the conductance. Increasing or decreasing the bias while having the peptide gate trapped in the pore stretches or compresses the peptide within the nanopore, thus modulating its conductance in a non-linear but reproducible manner. We envision a range of applications of this removable-gate nanopore system, e.g. from an element of biological computing circuits to a test bed for probing the elasticity of intrinsically disordered proteins.
Changes in the conformation of the FraC nanopore during the 80 ns equilibration simulation. The protein’s alpha carbon atoms were constrained for the first 65 ns of the MD trajectory. The protein is shown using a cartoon representation; the phosphorous atoms of the lipid bilayer are shown as orange spheres, the lipid tails are shown in grey lines, the electrolyte solution is not shown
MD simulation of an open FraC nanopore (blue cut-away surface) embedded in a DPhPC membrane (grey lines and orange spheres) and submerged in 1 M NaCl solution (pink and yellow spheres representing Na+and Cl-ions, respectively), water not shown. A -100 mV bias was applied to produce current of ions through the FraC nanopore. The animation illustrates a 24 ns fragment of the MD trajectory.
MD simulation of the dipolar peptide gate capture under a -1.2V starting from a stretched conformation. The peptide has the following amino acid sequence: EEEEEEEEEECGSGSGSKGSRRRRRRRRRR. The movie illustrates a 12 ns fragment of the MD trajectory. The peptide is shown using spheres colored according to the amino acid charge: blue, red and green indicate positively charged, negatively charged, and neutral amino acids, respectively.
MD simulation of the truncated peptide capture under a -1.2V. The peptide has the following amino acid sequence: CGSGSGSKGSRRRRRRRRRR. The movie illustrates a 12 ns MD trajectory. The peptide is shown using spheres colored according to the amino acid charge: blue and green indicate positively charged and neutral amino acids, respectively.
MD simulation of the dipolar peptide gate capture under a -1.2V starting from a hairpin conformation. The peptide has the following amino acid sequence: EEEEEEEEEECGSGSGSKGSRRRRRRRRRR. The movie illustrates a 12 ns fragment of the MD trajectory. The peptide is shown using spheres colored according to the amino acid charge: blue, red and green indicate positively charged, negatively charged, and neutral amino acids, respectively.