Stretching and controlled motion of single-stranded DNA in locally heated solid-state nanopores
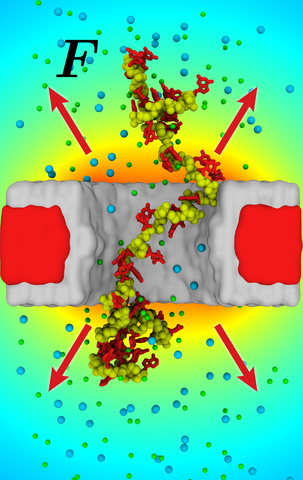
Practical applications of solid-state nanopores for DNA detection and sequencing require the electrophoretic motion of DNA through the nanopores to be precisely controlled. Controlling the motion of single-stranded DNA presents a particular challenge, in part because of the multitude of conformations that a DNA strand can adopt in a nanopore. Through continuum, coarse-grained and atomistic modeling, we demonstrate that local heating of the nanopore volume can be used to alter the electrophoretic mobility and conformation of single-stranded DNA. In the nanopore systems considered, the temperature near the nanopore is modulated via a nanometer-size heater element that can be radiatively switched on and off. The local enhancement of temperature produces considerable stretching of the DNA fragment confined within the nanopore. Such stretching is reversible, so that the conformation of DNA can be toggled between compact (local heating is off) and extended (local heating is on) states. The effective thermophoretic force acting on single-stranded DNA in the vicinity of the nanopore is found to be sufficiently large (4-8 pN) to affect such changes in the DNA conformation. The local heating of the nanopore volume is observed to promote single-file translocation of DNA strands at transmembrane biases as low as 10 mV, which opens new avenues for using solid-state nanopores for detection and sequencing of DNA.
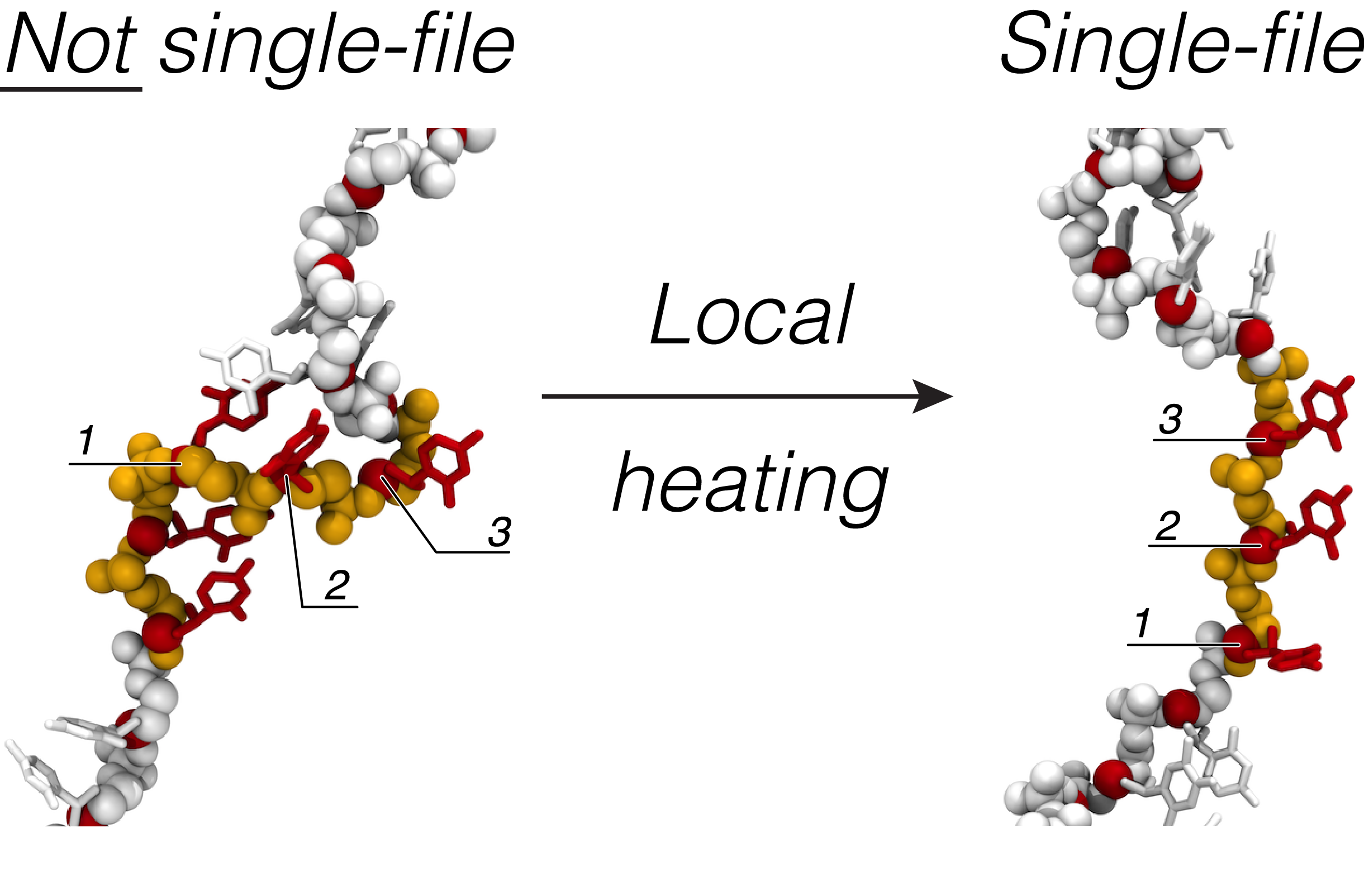
Unwinding of a single-stranded DNA in a locally heated Si3N4 nanopore. The Si3N4 membrane is shown in gray; the DNA molecule is shown in balls and sticks representation and colored to reflect its nucleotide sequence (blue adenine, yellow cytosine, red thymine and green guanine). The heated region of the membrane (shown in red) is kept at ΔT = 100 K higher than the ambient temperature, producing the temperature increase in the nanopore volume of 70 K. The animation shows 20 ns of all-atom MD simulation.
Relaxation of a single-stranded DNA molecule following the unwinding simulation. In this simulation, the heating source was switched off. The Si3N4 membrane is shown in gray; the DNA molecule is shown in balls and sticks representation and colored to reflect its nucleotide sequence (blue adenine, yellow cytosine, red thymine and green guanine). The animation covers 200 ns of all-atom MD simulation.
Coarse-grained MD simulation of a 250-nucleotide ssDNA fragment in a locally heated nanopore system. The local heater element temperature was set to be 100 K higher then the room temperature; the temperature of the solvent in the nanopore volume was ~ 70 K higher then the room temperature. One end of the DNA strand is restrained to remain in the center of the nanopore, the force of the restraint reports the effective force on the strand. The membrane is shown in gray, DNA molecule is shown in blue and white spheres for the backbone and base beads, correspondingly, and the restrained backbone atom is shown as red sphere. The animation covers 1.27 μs of CG MD simulation, which corresponds to ~ 12.7 μs of physical time.
Coarse-grained MD simulation of a 250-nucleotide ssDNA fragment in a locally heated nanopore system. No heating was applied in this simulation. One end of the DNA strand is restrained to the center of the nanopore, the force of the restrain reports the effective force on the strand. The membrane is shown in gray, DNA molecule is shown by blue and white spheres for the backbone and base beads, correspondingly, and the restrained backbone atom is shown as a red sphere. The animation covers 1.0 μs of CG MD simulation, which corresponds to ~ 10 μs of physical time.