Assessing graphene nanopores for sequencing DNA.
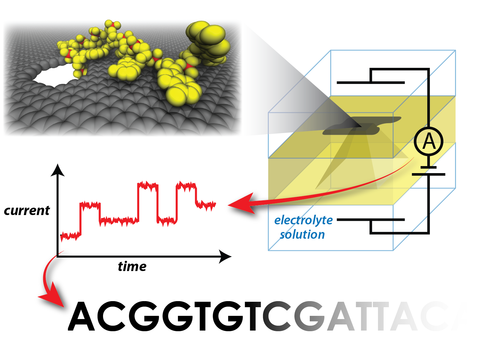
Using all-atom molecular dynamics and atomic-resolution Brownian dynamics, we simulate the translocation of single-stranded DNA through graphene nanopores and characterize the ionic current blockades produced by DNA nucleotides. We find that transport of single DNA strands through graphene nanopores may occur in single nucleotide steps. For certain pore geometries, hydrophobic interactions with the graphene membrane lead to a dramatic reduction in the conformational fluctuations of the nucleotides in the nanopores. Furthermore, we show that ionic current blockades produced by different DNA nucleotides are, in general, indicative of the nucleotide type, but very sensitive to the orientation of the nucleotides in the nanopore. Taken together, our simulations suggest that strand sequencing of DNA by measuring the ionic current blockades in graphene nanopores may be possible, given that the conformation of DNA nucleotides in the nanopore can be controlled through precise engineering of the nanopore surface.
A side and top-down view of a single-stranded DNA (ssDNA) molecule translocating through a three layer, free-standing graphene nanopore. Within the first 25 ns of simulation the ssDNA bases adsorb onto graphene. The adsorption is likely a hydrophobic effect as waters are excluded between the DNA bases and graphene. A 500 mV transmembrane bias drives the ssDNA through the nanopore.
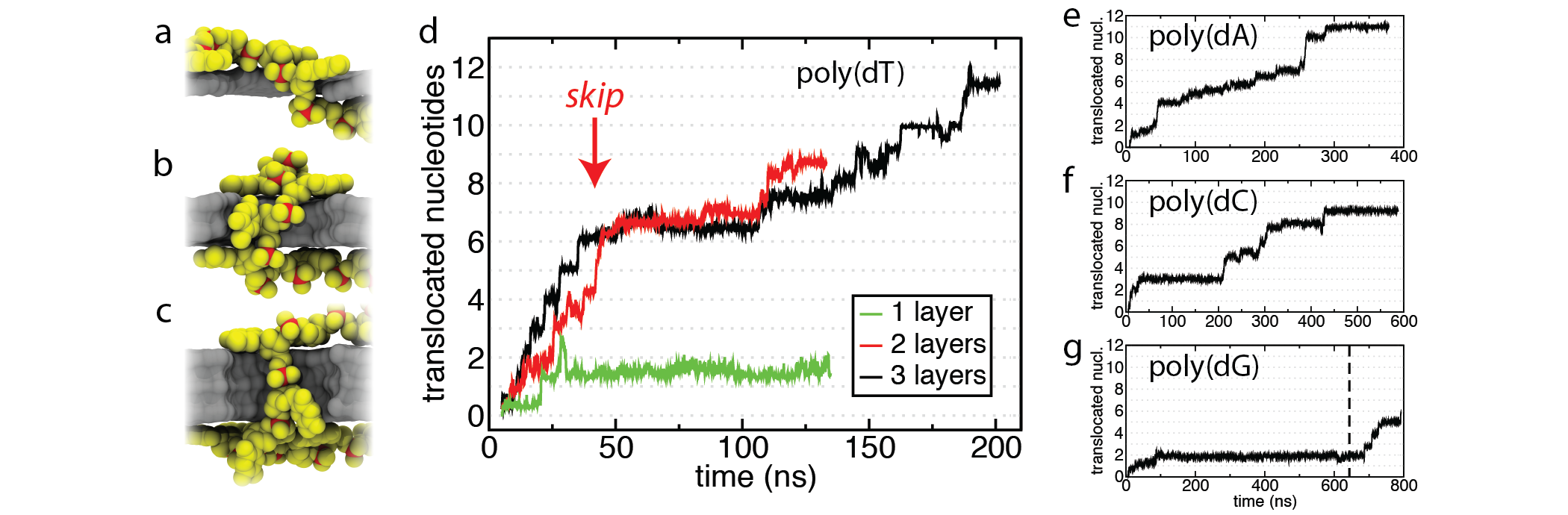
(a-c) Snapshots of DNA (yellow and red) inside 1, 2 and 3 layer graphene nanopores. We found that poly(dT)20 translocates without substantial skips through a nanopore in 3 layers of graphene (d, black) compared to 1 and 2 layers (green, red). Homopolyers of poly(dA)20, poly(dC)20, and poly(dG)20 also translocated through the three layer pore (e-g), although at much longer timescales compared to poly(dT)20.
 The movie above shows a top-down (left) and side view (right) of the single-stranded DNA molecule, displayed as alternating blue and orange nucleotides, moving through a nanopore in a three layer graphene membrane. Bases that are hydrophobically adhered to the top of the graphene membrane are driven into the nanopore by a 500mV transmembrane bias . The bases occupying the pore displace to the opposite side of the pore and promptly readsorb onto the bottom of the graphene membrane. The overall translocation process occurs in a step-wise manner as illustrated on the graph in the right.
The movie above shows a top-down (left) and side view (right) of the single-stranded DNA molecule, displayed as alternating blue and orange nucleotides, moving through a nanopore in a three layer graphene membrane. Bases that are hydrophobically adhered to the top of the graphene membrane are driven into the nanopore by a 500mV transmembrane bias . The bases occupying the pore displace to the opposite side of the pore and promptly readsorb onto the bottom of the graphene membrane. The overall translocation process occurs in a step-wise manner as illustrated on the graph in the right.
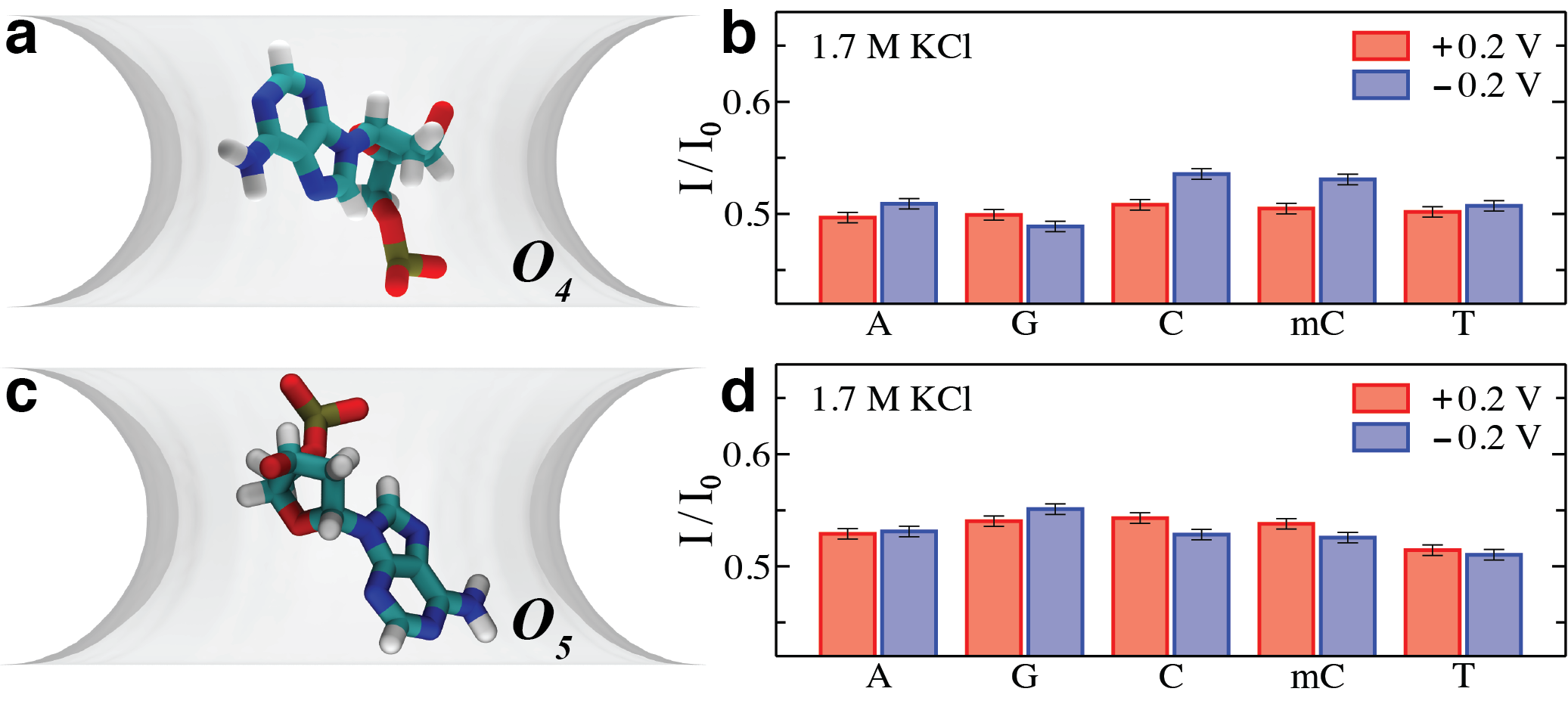
Changes in the ionic current provide a simple method of 'reading out' the DNA bases inside the nanopore in real-time. Here we used Brownian dynamics simulation to compute the ionic currents for each base in realistic conformations (a,c) inside the nanopore under a ±0.2 V transmembrane bias. Orientations (a,c) were taken as the most probable nucleotide orientations from MD simulations. In orientation 1 (a,b) cytosine and methylated cytosine can be distinguished under a -0.2 V bias. In orientation 2 (c) thymine can be recognized from other nucleotides under both a positive and negative biases.